Computer model of influenza virus shows universal vaccine promise
Each year there are an estimated 1 billion cases of influenza, including 3-5 million severe cases and up to 650,000 influenza-related respiratory deaths globally, according to the World Health Organization. Seasonal flu vaccines must be reformulated each year to match the predominantly circulating strains. When the vaccine matches the predominant strain, it is very effective; however, when it does not match, it may offer little protection.
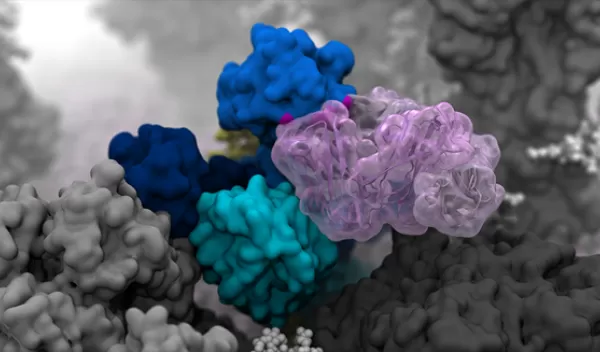
For the first time, U.S. National Science Foundation-supported researchers at the University of California San Diego have created an atomic-level computer model of the H1N1 virus that reveals new vulnerabilities through glycoprotein "breathing" and "tilting" movements. This work, published in ACS Central Science, suggests possible strategies for the design of future vaccines and antivirals against influenza.
The main targets of the flu vaccine are two surface glycoproteins, hemagglutinin, or HA, and neuraminidase, NA. While the HA protein helps the virus bind to the host cell, the NA protein acts like scissors to cut the HA away from the cell membrane, allowing the virus to replicate. Although the properties of both glycoproteins have been studied previously, a complete understanding of their movement does not exist.
"Once we knew our models were correct, we realized the enormous potential this discovery held," said Rommie Amaro, principal investigator on the project. "This research could be used to develop methods of keeping the protein locked open so that it would be constantly accessible to antibodies."
Amaro is making the data available to other researchers who can uncover even more about how the influenza virus moves, grows and evolves. "This paves the way for other groups to apply similar methods to other viruses," Amaro said. "We've modeled SARS-CoV-2 in the past and now H1N1, but there are other flu variants, MERS, RSV, HIV — this is just the beginning."
