Once Considered Too High-Risk, Supercomputer Simulations of 'Wiggling and Jiggling' Atoms Could Help Stop Coronavirus
UC San Diego Professor Rommie Amaro had an idea. The way it was, you took snapshots of a single protein to figure out its atomic structure. What Amaro wanted to do, use snapshots from many proteins to create a moving, all-atom replica of a virus.
The idea wasn't met with what she calls "grand reception." At least, not at first.
But with support from the National Science Foundation (NSF), through its supercomputing resources, Amaro's lab ran model simulations of the 2009 H1N1 virus (Swine Flu). It has chilling similarities to pathogenic foes past and present, to the H1N1 virus that caused the 1918 Spanish Flu Pandemic and to SARS-CoV-2—the 2019 novel coronavirus.
February 2020, the Amaro lab published research on breakthrough virus simulations. Now, in the race against the COVID-19 virus, Amaro is leading an international team to create a model on one of the most powerful computers in the world. Her idea could help reveal vulnerabilities. It could help resolve one of the darkest moments in modern history.
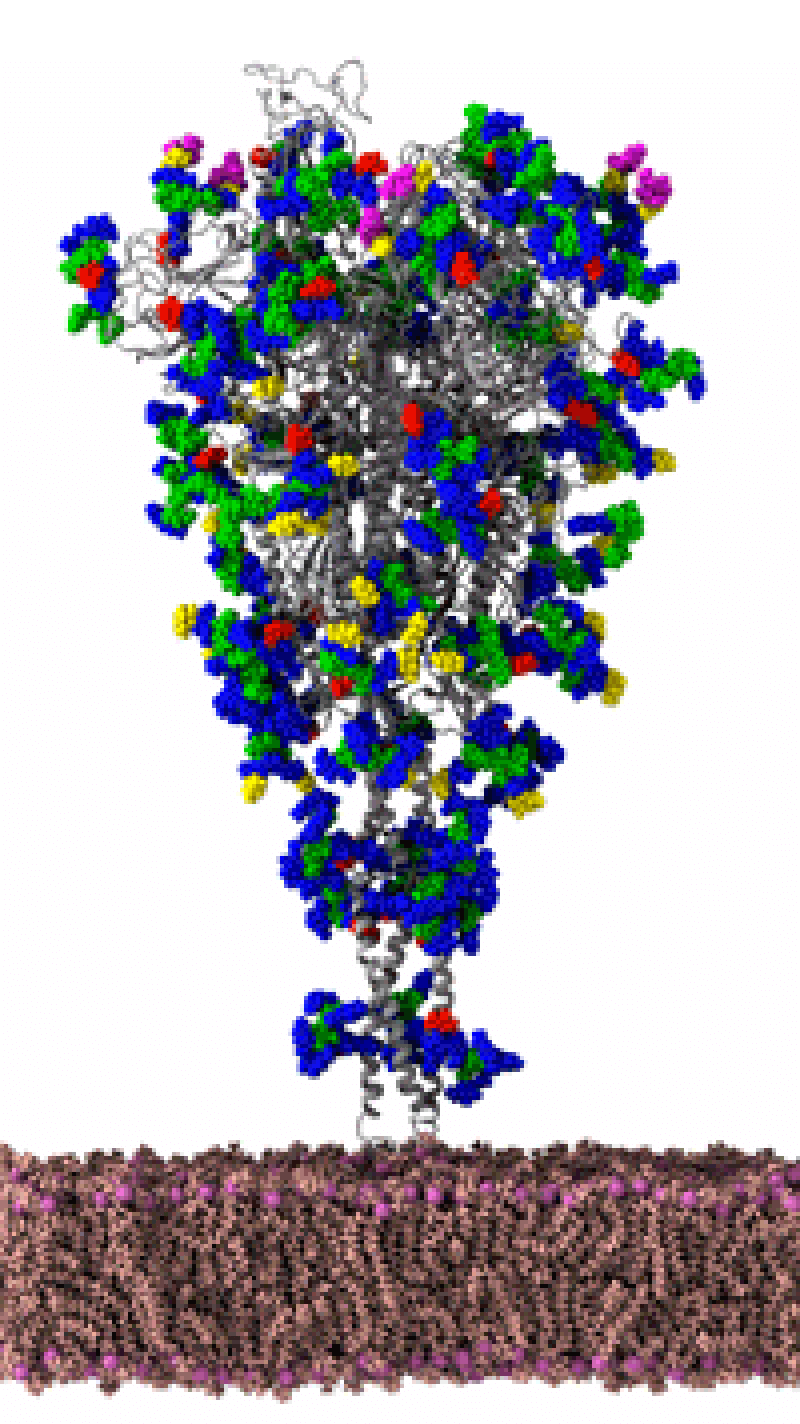
Specifically, the team is creating a moving simulation of the SARS-CoV-2 envelope. Modeling this exterior structure of the virus—fats, glycans, gnarly proteins—could reveal aspects of molecular recognition, how the virus interacts to infect a host cell. Here's why it can't be done on a typical computer: The model will include the envelope's atoms—all of them, some 200 million.
So, the team is using Frontera, the NSF-funded supercomputer launched last year at the Texas Advanced Computer Center. It's the fifth most powerful computer in the world; the fastest on any U.S. university campus. It can compute in one second what one person can calculate every second for about one billion years. Amaro's simulations are so intense, they're using about 250,000 processing cores—half of Frontera's computation capability.
Wiggling and Jiggling
Amaro describes it as a "computational microscope." Experimental imaging techniques, like cryo-electron microscopy and X-ray crystallography, offer snapshots of molecules on the atomic level. Amaro explains such techniques "do give us these beautiful images, the sort of Polaroid snapshots of these molecules." Amaro even has a few, framed, hanging in her office. But snapshots of a molecule don't capture molecular dynamics, how structures move. In that sense, Amaro's simulations are like gifs. They use experimental data to create moving models through molecular dynamics—"the wiggling and jiggling of atoms.”
Amaro uses the succinct description in reference to the famous quote by Physicist Richard Feynman, “Everything that living things do can be understood in terms of the jiggling and wiggling of atoms.” And knowing how viral machines wiggle and jiggle is important because it gives researchers a better chance to target vaccines and drugs. Recently, building on experimental work at UT Austin, Amaro's team created a moving visualization of a spiked-protein, the main protein involved in host-cell infection. Amaro shared the "movie" to Twitter, comparing the virus with the monster on Stranger Things.
It's a message full of hope: "#glycotime for the #COVID19 #demogorgon."
Credit: Amaro Lab
Risk
Amaro is an award-winning scientist, mother of four, triathlete, she once worked as an engineer at Kraft Foods designing Philadelphia Cream Cheese. She focused her early research career on diseases that science had "neglected." Top tags on her "Rate My Professor" profile: Caring, Respected, Amazing Lectures, Hilarious.
But in a recent talk, she recalled, "Whenever you are doing something that's sort of very different than what everybody else is doing—I think probably everybody in every field experiences this—it's very challenging." A great idea is not always recognized for what it is. "Because it's viewed as too high risk."
One reason NSF exists is to give the best high-risk ideas a chance. In 2014 and again in 2018, Amaro was granted NSF funding. She used Blue Waters—the NSF-funded predecessor to Frontera, located at the University of Illinois at Urbana-Champaign. She was able to visualize the H1N1 virus with a moving, all-atom super-computer simulation. The full 160 million atoms. "Probably the largest system ever simulated."
Amaro recalled that when she first wanted to create these super-computing simulations, "People were thinking, 'Well, what are you going to learn from this?'" Here's what: Within days of publishing research on the H1N1 virus, Amaro's team was working to model the COVID virus. The H1N1 virus doesn't have a capsid head. Neither does SARS-CoV-2. Both attach to a host through viral envelope. In other words, because of their research on H1N1, the Amaro team could respond quickly to the COVID-19 crisis—the H1N1 paper went to press on February 19, the team started working on SARS-CoV-2 nearly the same day.
Convergence
Though the work is urgent, Amaro recently shared a screenshot of a virtual meeting with a good-humored caption, "Amaro Lab's First Zooooommmmm." The team is international, the work is remote, Frontera is accessible on the cloud.
It is, at least, a model of intersecting and interdisciplinary science. It is an example of the power of converging science in scale and scope. There is convergence within the research enterprise, between NSF and the National Institute of Health, which support Amaro's research. There is convergence between institutes, among Amaro's UC lab and UT Austin. There is convergence in high performance computing and data science; in observational and simulation sciences; in chemistry, biology, physics and pharmacology. There is convergence in both applied and basic research.
This research is groundbreaking but, as Amaro states, matter of fact, "There will always be a next pandemic." And as long as there's a National Science Foundation, there will be capacity to respond through big ideas and high-risk research.
Written by Trisha Calvarese, Speechwriter in the Office of Legislative and Public Affairs, National Science Foundation / Prime Meridian Media Inc.
COVID-19 Resources: Coronavirus.gov; Coronavirus Disease 2019 (COVID-19); What the U.S. Government is Doing